Fitting with ScikitLearn - Part 2
Contents
Fitting with ScikitLearn - Part 2#
Overview
Questions
- How can I use scikitlearn to fit machine learning models?
Objectives:
- Slice a pandas dataframe to get `X` and `Y` values and convert them to NumPy Arrays.
- Use the `LinearRegression` model in scikitlearn to perform a linear fit.
Keypoints:
- You must import and create the model you want to use from scikitlearn.
- SciKitLearn models require `X` and `Y` values that are at least two dimensional.
- Use `.reshape` on your NumPy arrays to make sure they are the correct dimension.
- Fit SciKitLearn models by giving them data and using the `fit` method.
- Use the `predict` method after fitting to make predictions
Data Preparation#
import os
import pandas as pd
plates = pd.read_csv("data/rxnpredict/data_table.csv")
plates.head()
| plate | row | col | base | base_cas_number | base_smiles | ligand | ligand_cas_number | ligand_smiles | aryl_halide_number | aryl_halide | aryl_halide_smiles | additive_number | additive | additive_smiles | product_smiles | yield | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1 | 1 | P2Et | 165535-45-5 | CN(C)P(N(C)C)(N(C)C)=NP(N(C)C)(N(C)C)=NCC | XPhos | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 1.0 | 1-chloro-4-(trifluoromethyl)benzene | FC(F)(F)c1ccc(Cl)cc1 | NaN | NaN | NaN | Cc1ccc(Nc2ccc(C(F)(F)F)cc2)cc1 | 26.888615 |
| 1 | 1 | 1 | 2 | P2Et | 165535-45-5 | CN(C)P(N(C)C)(N(C)C)=NP(N(C)C)(N(C)C)=NCC | XPhos | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 2.0 | 1-bromo-4-(trifluoromethyl)benzene | FC(F)(F)c1ccc(Br)cc1 | NaN | NaN | NaN | Cc1ccc(Nc2ccc(C(F)(F)F)cc2)cc1 | 24.063224 |
| 2 | 1 | 1 | 3 | P2Et | 165535-45-5 | CN(C)P(N(C)C)(N(C)C)=NP(N(C)C)(N(C)C)=NCC | XPhos | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 3.0 | 1-iodo-4-(trifluoromethyl)benzene | FC(F)(F)c1ccc(I)cc1 | NaN | NaN | NaN | Cc1ccc(Nc2ccc(C(F)(F)F)cc2)cc1 | 47.515821 |
| 3 | 1 | 1 | 4 | P2Et | 165535-45-5 | CN(C)P(N(C)C)(N(C)C)=NP(N(C)C)(N(C)C)=NCC | XPhos | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 4.0 | 1-chloro-4-methoxybenzene | COc1ccc(Cl)cc1 | NaN | NaN | NaN | COc1ccc(Nc2ccc(C)cc2)cc1 | 2.126831 |
| 4 | 1 | 1 | 5 | P2Et | 165535-45-5 | CN(C)P(N(C)C)(N(C)C)=NP(N(C)C)(N(C)C)=NCC | XPhos | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 5.0 | 1-bromo-4-methoxybenzene | COc1ccc(Br)cc1 | NaN | NaN | NaN | COc1ccc(Nc2ccc(C)cc2)cc1 | 47.586354 |
descriptors = pd.read_csv("data/rxnpredict/output_table_modified.csv")
descriptors.head()
| additive_*C3_NMR_shift | additive_*C3_electrostatic_charge | additive_*C4_NMR_shift | additive_*C4_electrostatic_charge | additive_*C5_NMR_shift | additive_*C5_electrostatic_charge | additive_*N1_electrostatic_charge | additive_*O1_electrostatic_charge | additive_E_HOMO | additive_E_LUMO | ... | ligand_V6_intensity | ligand_V7_frequency | ligand_V7_intensity | ligand_V8_frequency | ligand_V8_intensity | ligand_V9_frequency | ligand_V9_intensity | ligand_dipole_moment | plate | row | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 4.414 | 3026.561 | 16.577 | 3043.097 | 18.145 | 3064.344 | 38.21 | 1.212924 | 1 | 1 |
| 1 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 4.414 | 3026.561 | 16.577 | 3043.097 | 18.145 | 3064.344 | 38.21 | 1.212924 | 1 | 1 |
| 2 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 4.414 | 3026.561 | 16.577 | 3043.097 | 18.145 | 3064.344 | 38.21 | 1.212924 | 1 | 1 |
| 3 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 4.414 | 3026.561 | 16.577 | 3043.097 | 18.145 | 3064.344 | 38.21 | 1.212924 | 1 | 1 |
| 4 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 4.414 | 3026.561 | 16.577 | 3043.097 | 18.145 | 3064.344 | 38.21 | 1.212924 | 1 | 1 |
5 rows × 123 columns
# Join on common columns
# Pull out just what we need
plates_join = plates[["plate", "row", "col", "yield"]]
dataset = pd.merge(descriptors, plates, on=["plate", "row", "col"])
dataset.head()
| additive_*C3_NMR_shift | additive_*C3_electrostatic_charge | additive_*C4_NMR_shift | additive_*C4_electrostatic_charge | additive_*C5_NMR_shift | additive_*C5_electrostatic_charge | additive_*N1_electrostatic_charge | additive_*O1_electrostatic_charge | additive_E_HOMO | additive_E_LUMO | ... | ligand_cas_number | ligand_smiles | aryl_halide_number | aryl_halide | aryl_halide_smiles | additive_number | additive | additive_smiles | product_smiles | yield | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 1.0 | 1-chloro-4-(trifluoromethyl)benzene | FC(F)(F)c1ccc(Cl)cc1 | NaN | NaN | NaN | Cc1ccc(Nc2ccc(C(F)(F)F)cc2)cc1 | 26.888615 |
| 1 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 2.0 | 1-bromo-4-(trifluoromethyl)benzene | FC(F)(F)c1ccc(Br)cc1 | NaN | NaN | NaN | Cc1ccc(Nc2ccc(C(F)(F)F)cc2)cc1 | 24.063224 |
| 2 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 3.0 | 1-iodo-4-(trifluoromethyl)benzene | FC(F)(F)c1ccc(I)cc1 | NaN | NaN | NaN | Cc1ccc(Nc2ccc(C(F)(F)F)cc2)cc1 | 47.515821 |
| 3 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 4.0 | 1-chloro-4-methoxybenzene | COc1ccc(Cl)cc1 | NaN | NaN | NaN | COc1ccc(Nc2ccc(C)cc2)cc1 | 2.126831 |
| 4 | 143.12 | 0.223 | 93.06 | -0.447 | 162.34 | 0.292 | -0.334 | -0.057 | -0.2317 | -0.0487 | ... | 564483-18-7 | CC(C)C1=CC(C(C)C)=CC(C(C)C)=C1C2=C(P(C3CCCCC3)... | 5.0 | 1-bromo-4-methoxybenzene | COc1ccc(Br)cc1 | NaN | NaN | NaN | COc1ccc(Nc2ccc(C)cc2)cc1 | 47.586354 |
5 rows × 137 columns
Using SciKitLearn to Fit#
X = dataset[descriptors.columns].to_numpy()
Y = dataset["yield"].to_numpy()
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_fit = sc.fit_transform(X)
from sklearn.ensemble import RandomForestRegressor
help(RandomForestRegressor)
Help on class RandomForestRegressor in module sklearn.ensemble._forest:
class RandomForestRegressor(ForestRegressor)
| RandomForestRegressor(n_estimators=100, *, criterion='mse', max_depth=None, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features='auto', max_leaf_nodes=None, min_impurity_decrease=0.0, min_impurity_split=None, bootstrap=True, oob_score=False, n_jobs=None, random_state=None, verbose=0, warm_start=False, ccp_alpha=0.0, max_samples=None)
|
| A random forest regressor.
|
| A random forest is a meta estimator that fits a number of classifying
| decision trees on various sub-samples of the dataset and uses averaging
| to improve the predictive accuracy and control over-fitting.
| The sub-sample size is controlled with the `max_samples` parameter if
| `bootstrap=True` (default), otherwise the whole dataset is used to build
| each tree.
|
| Read more in the :ref:`User Guide <forest>`.
|
| Parameters
| ----------
| n_estimators : int, default=100
| The number of trees in the forest.
|
| .. versionchanged:: 0.22
| The default value of ``n_estimators`` changed from 10 to 100
| in 0.22.
|
| criterion : {"mse", "mae"}, default="mse"
| The function to measure the quality of a split. Supported criteria
| are "mse" for the mean squared error, which is equal to variance
| reduction as feature selection criterion, and "mae" for the mean
| absolute error.
|
| .. versionadded:: 0.18
| Mean Absolute Error (MAE) criterion.
|
| max_depth : int, default=None
| The maximum depth of the tree. If None, then nodes are expanded until
| all leaves are pure or until all leaves contain less than
| min_samples_split samples.
|
| min_samples_split : int or float, default=2
| The minimum number of samples required to split an internal node:
|
| - If int, then consider `min_samples_split` as the minimum number.
| - If float, then `min_samples_split` is a fraction and
| `ceil(min_samples_split * n_samples)` are the minimum
| number of samples for each split.
|
| .. versionchanged:: 0.18
| Added float values for fractions.
|
| min_samples_leaf : int or float, default=1
| The minimum number of samples required to be at a leaf node.
| A split point at any depth will only be considered if it leaves at
| least ``min_samples_leaf`` training samples in each of the left and
| right branches. This may have the effect of smoothing the model,
| especially in regression.
|
| - If int, then consider `min_samples_leaf` as the minimum number.
| - If float, then `min_samples_leaf` is a fraction and
| `ceil(min_samples_leaf * n_samples)` are the minimum
| number of samples for each node.
|
| .. versionchanged:: 0.18
| Added float values for fractions.
|
| min_weight_fraction_leaf : float, default=0.0
| The minimum weighted fraction of the sum total of weights (of all
| the input samples) required to be at a leaf node. Samples have
| equal weight when sample_weight is not provided.
|
| max_features : {"auto", "sqrt", "log2"}, int or float, default="auto"
| The number of features to consider when looking for the best split:
|
| - If int, then consider `max_features` features at each split.
| - If float, then `max_features` is a fraction and
| `round(max_features * n_features)` features are considered at each
| split.
| - If "auto", then `max_features=n_features`.
| - If "sqrt", then `max_features=sqrt(n_features)`.
| - If "log2", then `max_features=log2(n_features)`.
| - If None, then `max_features=n_features`.
|
| Note: the search for a split does not stop until at least one
| valid partition of the node samples is found, even if it requires to
| effectively inspect more than ``max_features`` features.
|
| max_leaf_nodes : int, default=None
| Grow trees with ``max_leaf_nodes`` in best-first fashion.
| Best nodes are defined as relative reduction in impurity.
| If None then unlimited number of leaf nodes.
|
| min_impurity_decrease : float, default=0.0
| A node will be split if this split induces a decrease of the impurity
| greater than or equal to this value.
|
| The weighted impurity decrease equation is the following::
|
| N_t / N * (impurity - N_t_R / N_t * right_impurity
| - N_t_L / N_t * left_impurity)
|
| where ``N`` is the total number of samples, ``N_t`` is the number of
| samples at the current node, ``N_t_L`` is the number of samples in the
| left child, and ``N_t_R`` is the number of samples in the right child.
|
| ``N``, ``N_t``, ``N_t_R`` and ``N_t_L`` all refer to the weighted sum,
| if ``sample_weight`` is passed.
|
| .. versionadded:: 0.19
|
| min_impurity_split : float, default=None
| Threshold for early stopping in tree growth. A node will split
| if its impurity is above the threshold, otherwise it is a leaf.
|
| .. deprecated:: 0.19
| ``min_impurity_split`` has been deprecated in favor of
| ``min_impurity_decrease`` in 0.19. The default value of
| ``min_impurity_split`` has changed from 1e-7 to 0 in 0.23 and it
| will be removed in 1.0 (renaming of 0.25).
| Use ``min_impurity_decrease`` instead.
|
| bootstrap : bool, default=True
| Whether bootstrap samples are used when building trees. If False, the
| whole dataset is used to build each tree.
|
| oob_score : bool, default=False
| Whether to use out-of-bag samples to estimate the generalization score.
| Only available if bootstrap=True.
|
| n_jobs : int, default=None
| The number of jobs to run in parallel. :meth:`fit`, :meth:`predict`,
| :meth:`decision_path` and :meth:`apply` are all parallelized over the
| trees. ``None`` means 1 unless in a :obj:`joblib.parallel_backend`
| context. ``-1`` means using all processors. See :term:`Glossary
| <n_jobs>` for more details.
|
| random_state : int, RandomState instance or None, default=None
| Controls both the randomness of the bootstrapping of the samples used
| when building trees (if ``bootstrap=True``) and the sampling of the
| features to consider when looking for the best split at each node
| (if ``max_features < n_features``).
| See :term:`Glossary <random_state>` for details.
|
| verbose : int, default=0
| Controls the verbosity when fitting and predicting.
|
| warm_start : bool, default=False
| When set to ``True``, reuse the solution of the previous call to fit
| and add more estimators to the ensemble, otherwise, just fit a whole
| new forest. See :term:`the Glossary <warm_start>`.
|
| ccp_alpha : non-negative float, default=0.0
| Complexity parameter used for Minimal Cost-Complexity Pruning. The
| subtree with the largest cost complexity that is smaller than
| ``ccp_alpha`` will be chosen. By default, no pruning is performed. See
| :ref:`minimal_cost_complexity_pruning` for details.
|
| .. versionadded:: 0.22
|
| max_samples : int or float, default=None
| If bootstrap is True, the number of samples to draw from X
| to train each base estimator.
|
| - If None (default), then draw `X.shape[0]` samples.
| - If int, then draw `max_samples` samples.
| - If float, then draw `max_samples * X.shape[0]` samples. Thus,
| `max_samples` should be in the interval `(0, 1)`.
|
| .. versionadded:: 0.22
|
| Attributes
| ----------
| base_estimator_ : DecisionTreeRegressor
| The child estimator template used to create the collection of fitted
| sub-estimators.
|
| estimators_ : list of DecisionTreeRegressor
| The collection of fitted sub-estimators.
|
| feature_importances_ : ndarray of shape (n_features,)
| The impurity-based feature importances.
| The higher, the more important the feature.
| The importance of a feature is computed as the (normalized)
| total reduction of the criterion brought by that feature. It is also
| known as the Gini importance.
|
| Warning: impurity-based feature importances can be misleading for
| high cardinality features (many unique values). See
| :func:`sklearn.inspection.permutation_importance` as an alternative.
|
| n_features_ : int
| The number of features when ``fit`` is performed.
|
| n_outputs_ : int
| The number of outputs when ``fit`` is performed.
|
| oob_score_ : float
| Score of the training dataset obtained using an out-of-bag estimate.
| This attribute exists only when ``oob_score`` is True.
|
| oob_prediction_ : ndarray of shape (n_samples,)
| Prediction computed with out-of-bag estimate on the training set.
| This attribute exists only when ``oob_score`` is True.
|
| See Also
| --------
| DecisionTreeRegressor, ExtraTreesRegressor
|
| Notes
| -----
| The default values for the parameters controlling the size of the trees
| (e.g. ``max_depth``, ``min_samples_leaf``, etc.) lead to fully grown and
| unpruned trees which can potentially be very large on some data sets. To
| reduce memory consumption, the complexity and size of the trees should be
| controlled by setting those parameter values.
|
| The features are always randomly permuted at each split. Therefore,
| the best found split may vary, even with the same training data,
| ``max_features=n_features`` and ``bootstrap=False``, if the improvement
| of the criterion is identical for several splits enumerated during the
| search of the best split. To obtain a deterministic behaviour during
| fitting, ``random_state`` has to be fixed.
|
| The default value ``max_features="auto"`` uses ``n_features``
| rather than ``n_features / 3``. The latter was originally suggested in
| [1], whereas the former was more recently justified empirically in [2].
|
| References
| ----------
| .. [1] L. Breiman, "Random Forests", Machine Learning, 45(1), 5-32, 2001.
|
| .. [2] P. Geurts, D. Ernst., and L. Wehenkel, "Extremely randomized
| trees", Machine Learning, 63(1), 3-42, 2006.
|
| Examples
| --------
| >>> from sklearn.ensemble import RandomForestRegressor
| >>> from sklearn.datasets import make_regression
| >>> X, y = make_regression(n_features=4, n_informative=2,
| ... random_state=0, shuffle=False)
| >>> regr = RandomForestRegressor(max_depth=2, random_state=0)
| >>> regr.fit(X, y)
| RandomForestRegressor(...)
| >>> print(regr.predict([[0, 0, 0, 0]]))
| [-8.32987858]
|
| Method resolution order:
| RandomForestRegressor
| ForestRegressor
| sklearn.base.RegressorMixin
| BaseForest
| sklearn.base.MultiOutputMixin
| sklearn.ensemble._base.BaseEnsemble
| sklearn.base.MetaEstimatorMixin
| sklearn.base.BaseEstimator
| builtins.object
|
| Methods defined here:
|
| __init__(self, n_estimators=100, *, criterion='mse', max_depth=None, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features='auto', max_leaf_nodes=None, min_impurity_decrease=0.0, min_impurity_split=None, bootstrap=True, oob_score=False, n_jobs=None, random_state=None, verbose=0, warm_start=False, ccp_alpha=0.0, max_samples=None)
| Initialize self. See help(type(self)) for accurate signature.
|
| ----------------------------------------------------------------------
| Data and other attributes defined here:
|
| __abstractmethods__ = frozenset()
|
| ----------------------------------------------------------------------
| Methods inherited from ForestRegressor:
|
| predict(self, X)
| Predict regression target for X.
|
| The predicted regression target of an input sample is computed as the
| mean predicted regression targets of the trees in the forest.
|
| Parameters
| ----------
| X : {array-like, sparse matrix} of shape (n_samples, n_features)
| The input samples. Internally, its dtype will be converted to
| ``dtype=np.float32``. If a sparse matrix is provided, it will be
| converted into a sparse ``csr_matrix``.
|
| Returns
| -------
| y : ndarray of shape (n_samples,) or (n_samples, n_outputs)
| The predicted values.
|
| ----------------------------------------------------------------------
| Methods inherited from sklearn.base.RegressorMixin:
|
| score(self, X, y, sample_weight=None)
| Return the coefficient of determination :math:`R^2` of the
| prediction.
|
| The coefficient :math:`R^2` is defined as :math:`(1 - \frac{u}{v})`,
| where :math:`u` is the residual sum of squares ``((y_true - y_pred)
| ** 2).sum()`` and :math:`v` is the total sum of squares ``((y_true -
| y_true.mean()) ** 2).sum()``. The best possible score is 1.0 and it
| can be negative (because the model can be arbitrarily worse). A
| constant model that always predicts the expected value of `y`,
| disregarding the input features, would get a :math:`R^2` score of
| 0.0.
|
| Parameters
| ----------
| X : array-like of shape (n_samples, n_features)
| Test samples. For some estimators this may be a precomputed
| kernel matrix or a list of generic objects instead with shape
| ``(n_samples, n_samples_fitted)``, where ``n_samples_fitted``
| is the number of samples used in the fitting for the estimator.
|
| y : array-like of shape (n_samples,) or (n_samples, n_outputs)
| True values for `X`.
|
| sample_weight : array-like of shape (n_samples,), default=None
| Sample weights.
|
| Returns
| -------
| score : float
| :math:`R^2` of ``self.predict(X)`` wrt. `y`.
|
| Notes
| -----
| The :math:`R^2` score used when calling ``score`` on a regressor uses
| ``multioutput='uniform_average'`` from version 0.23 to keep consistent
| with default value of :func:`~sklearn.metrics.r2_score`.
| This influences the ``score`` method of all the multioutput
| regressors (except for
| :class:`~sklearn.multioutput.MultiOutputRegressor`).
|
| ----------------------------------------------------------------------
| Data descriptors inherited from sklearn.base.RegressorMixin:
|
| __dict__
| dictionary for instance variables (if defined)
|
| __weakref__
| list of weak references to the object (if defined)
|
| ----------------------------------------------------------------------
| Methods inherited from BaseForest:
|
| apply(self, X)
| Apply trees in the forest to X, return leaf indices.
|
| Parameters
| ----------
| X : {array-like, sparse matrix} of shape (n_samples, n_features)
| The input samples. Internally, its dtype will be converted to
| ``dtype=np.float32``. If a sparse matrix is provided, it will be
| converted into a sparse ``csr_matrix``.
|
| Returns
| -------
| X_leaves : ndarray of shape (n_samples, n_estimators)
| For each datapoint x in X and for each tree in the forest,
| return the index of the leaf x ends up in.
|
| decision_path(self, X)
| Return the decision path in the forest.
|
| .. versionadded:: 0.18
|
| Parameters
| ----------
| X : {array-like, sparse matrix} of shape (n_samples, n_features)
| The input samples. Internally, its dtype will be converted to
| ``dtype=np.float32``. If a sparse matrix is provided, it will be
| converted into a sparse ``csr_matrix``.
|
| Returns
| -------
| indicator : sparse matrix of shape (n_samples, n_nodes)
| Return a node indicator matrix where non zero elements indicates
| that the samples goes through the nodes. The matrix is of CSR
| format.
|
| n_nodes_ptr : ndarray of shape (n_estimators + 1,)
| The columns from indicator[n_nodes_ptr[i]:n_nodes_ptr[i+1]]
| gives the indicator value for the i-th estimator.
|
| fit(self, X, y, sample_weight=None)
| Build a forest of trees from the training set (X, y).
|
| Parameters
| ----------
| X : {array-like, sparse matrix} of shape (n_samples, n_features)
| The training input samples. Internally, its dtype will be converted
| to ``dtype=np.float32``. If a sparse matrix is provided, it will be
| converted into a sparse ``csc_matrix``.
|
| y : array-like of shape (n_samples,) or (n_samples, n_outputs)
| The target values (class labels in classification, real numbers in
| regression).
|
| sample_weight : array-like of shape (n_samples,), default=None
| Sample weights. If None, then samples are equally weighted. Splits
| that would create child nodes with net zero or negative weight are
| ignored while searching for a split in each node. In the case of
| classification, splits are also ignored if they would result in any
| single class carrying a negative weight in either child node.
|
| Returns
| -------
| self : object
|
| ----------------------------------------------------------------------
| Readonly properties inherited from BaseForest:
|
| feature_importances_
| The impurity-based feature importances.
|
| The higher, the more important the feature.
| The importance of a feature is computed as the (normalized)
| total reduction of the criterion brought by that feature. It is also
| known as the Gini importance.
|
| Warning: impurity-based feature importances can be misleading for
| high cardinality features (many unique values). See
| :func:`sklearn.inspection.permutation_importance` as an alternative.
|
| Returns
| -------
| feature_importances_ : ndarray of shape (n_features,)
| The values of this array sum to 1, unless all trees are single node
| trees consisting of only the root node, in which case it will be an
| array of zeros.
|
| ----------------------------------------------------------------------
| Methods inherited from sklearn.ensemble._base.BaseEnsemble:
|
| __getitem__(self, index)
| Return the index'th estimator in the ensemble.
|
| __iter__(self)
| Return iterator over estimators in the ensemble.
|
| __len__(self)
| Return the number of estimators in the ensemble.
|
| ----------------------------------------------------------------------
| Data and other attributes inherited from sklearn.ensemble._base.BaseEnsemble:
|
| __annotations__ = {'_required_parameters': typing.List[str]}
|
| ----------------------------------------------------------------------
| Methods inherited from sklearn.base.BaseEstimator:
|
| __getstate__(self)
|
| __repr__(self, N_CHAR_MAX=700)
| Return repr(self).
|
| __setstate__(self, state)
|
| get_params(self, deep=True)
| Get parameters for this estimator.
|
| Parameters
| ----------
| deep : bool, default=True
| If True, will return the parameters for this estimator and
| contained subobjects that are estimators.
|
| Returns
| -------
| params : dict
| Parameter names mapped to their values.
|
| set_params(self, **params)
| Set the parameters of this estimator.
|
| The method works on simple estimators as well as on nested objects
| (such as :class:`~sklearn.pipeline.Pipeline`). The latter have
| parameters of the form ``<component>__<parameter>`` so that it's
| possible to update each component of a nested object.
|
| Parameters
| ----------
| **params : dict
| Estimator parameters.
|
| Returns
| -------
| self : estimator instance
| Estimator instance.
regressor = RandomForestRegressor()
regressor.fit(X_fit, Y)
RandomForestRegressor()
y_pred = regressor.predict(X_fit)
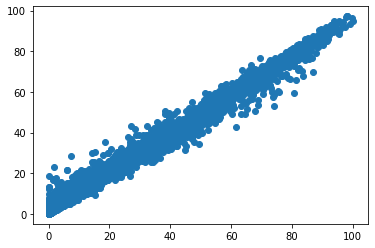
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.scatter(Y, y_pred)
<matplotlib.collections.PathCollection at 0x7facb9a576a0>
Training Testing Split#
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
X_train, X_test, Y_train, Y_test = train_test_split(X_fit, Y, test_size=0.30)
regressor = RandomForestRegressor()
regressor.fit(X_train, Y_train)
RandomForestRegressor()
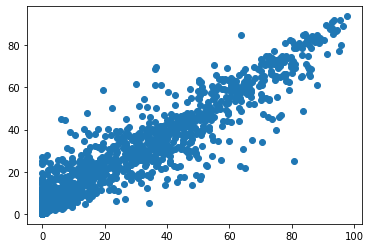
y_pred = regressor.predict(X_test)
plt.scatter(Y_test, y_pred)
<matplotlib.collections.PathCollection at 0x7facb18844f0>
from sklearn import metrics
import math
metrics.r2_score(Y_test, y_pred)
0.8545731030213027
math.sqrt(metrics.mean_squared_error(Y_test, y_pred))
9.966163336163948

